Research conducted at LCPP
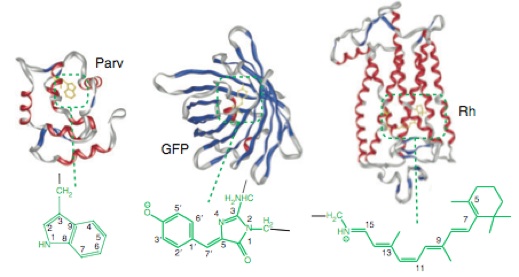
Current research at LCPP is centered on the idea that the most efficient and yet tunable photochemical systems know in nature are biological photoreceptors (i.e. visual and sensory pigments and ion-pumps). For this reason two parallel research lines are pursued. The first line focuses on the investigation of the molecular mechanism making a photoreceptor (a protein) or its mutants efficient in terms of color tuning (both in absorption and emission), reaction selectivity, time scale and quantum yield.
The second and parallel research line uses the same computational tools to design novel synthetic molecules that mimic the behavior of biological photoreceptors and can be employed as bio-mimetic molecular devices. For instance,
one of the LCPP targets is the development of a library of photo-responsive unnatural amino-acids to be employed in biological or medical research. Due to its research and the need for efficient computer programs and hardware, LCPP constitutes a potential environment for interdisciplinary work involving the Computer Sciences, Biology and Chemistry including organic synthesis.
To pursue the above targets, LCPP uses innovative computational protocols. These protocols, whose efficiency in treating large molecular systems has been demonstrated, aim to the simulation of the behavior of an entire population of photo-excited molecules thus overcoming the limitations of ordinary single molecule simulations. While, presently, this represents a brute-force computational effort, such development is foreseen to dominate the computer aided design of photoresponsive material in the near future. We believe that this is apparent when considering that: (i) the majority of the technological application of photo responsive materials involve the coherent (laser) or incoherent light irradiation of an entire sample and (ii) the availability of larger and larger arrays of fast processors (i.e. of computer clusters) will enable the simultaneous modeling of a statistically meaningful number of excited state molecules and, in turn, of an excited state population.
Research Projects at LCPP include:
•Photoisomerization Dynamics in Protein Cavities
•Bio-mimetic and Photochemical Switches and Motors
•Photocromic Microbial Rhodopsins
•Artificial Rhodopsin
Facilities
LCPP is a bi-national Lab which exploits the computer facilities both at the Department of Biotechnology, Chemistry and Pharmacy, University of Siena (UNISI), Italy, and at the Center for Photochemical Sciences, Department of Chemistry, Bowling Green State University (BGSU), Ohio. The laboratory, equipped with a pool of Linux-based servers including a 26 nodes – 134 cores High Performance Computer Cluster based on Intel Xeon (Woodcrest and Clovertown) CPUs and installed at the BGSU campus. The currently most advanced software tool used in the laboratory is a quantum-mechanics/molecular-mechanics software based on multiconfigurational quantum chemistry.
This facilities allow for the construction of realistic (i.e. quantitative) computer models of photo-excited chemical and biological systems. Most important LCPP aims at the creation of specific tools capable to track the time-evolution of the system making the lab activity highly complementary to that of the Ohio Laboratory of Kinetic and Spectrometry to which the LCPP has access. A larger number of processors and an improved software performance are being pursued to allow for a routinely affordable simulation time of excited state molecular population dynamics.


